New Publication in Nucleic Acid Research
INDEL detection, the ‘Achilles heel’ of precise genome editing: a survey of methods for accurate profiling of gene editing induced indels
Authors: Eric Paul Bennett, Bent Larsen Petersen, Ida Elisabeth Johansen, Yiyuan Niu, Zhang Yang, Christopher Aled Chamberlain, Özcan Met, Hans H. Wandall, Morten Frödin
Abstract:
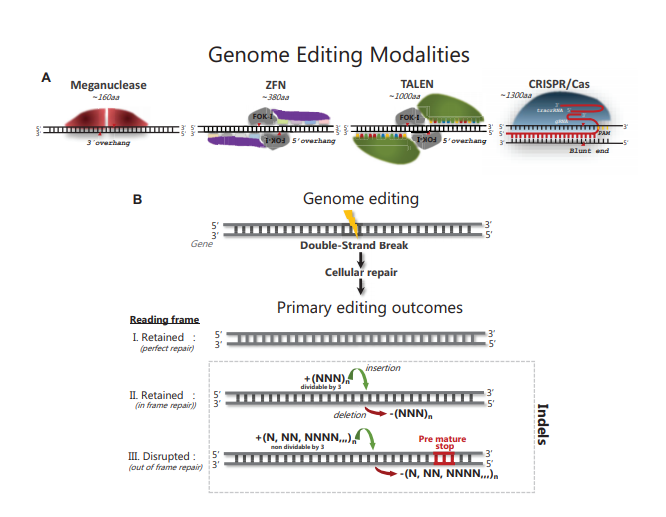
Advances in genome editing technologies have enabled manipulation of genomes at the single base level. These technologies are based on programmable nucleases (PNs) that include meganucleases, zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated 9 (Cas9) nucleases and have given researchers the ability to delete, insert or replace genomic DNA in cells, tissues and whole organisms. The great flexibility in re-designing the genomic target specificity of PNs has vastly expanded the scope of gene editing applications in life science, and shows great promise for development of the next generation gene therapies. PN technologies share the principle of inducing a DNA double-strand break (DSB) at a user-specified site in the genome, followed by cellular repair of the induced DSB. PN-elicited DSBs are mainly repaired by the non-homologous end joining (NHEJ) and the microhomology-mediated end joining (MMEJ) pathways, which can elicit a variety of small insertion or deletion (indel) mutations. If indels are elicited in a protein coding sequence and shift the reading frame, targeted gene knock out (KO) can readily be achieved using either of the available PNs. Despite the ease by which gene inactivation in principle can be achieved, in practice, successful KO is not only determined by the efficiency of NHEJ and MMEJ repair; it also depends on the design and properties of the PN utilized, delivery format chosen, the preferred indel repair outcomes at the targeted site, the chromatin state of the target site and the relative activities of the repair pathways in the edited cells. These variables preclude accurate prediction of the nature and frequency of PN induced indels. A key step of any gene KO experiment therefore becomes the detection, characterization and quantification of the indel(s) induced at the targeted genomic site in cells, tissues or whole organisms. In this survey, we briefly review naturally occurring indels and their detection. Next, we review the methods that have been developed for detection of PN-induced indels. We briefly outline the experimental steps and describe the pros and cons of the various methods to help users decide a suitable method for their editing application. We highlight recent advances that enable accurate and sensitive quantification of indel events in cells regardless of their genome complexity, turning a complex pool of different indel events into informative indel profiles. Finally, we review what has been learned about PN-elicited indel formation through the use of the new methods and how this insight is helping to further advance the genome editing field.