Resources
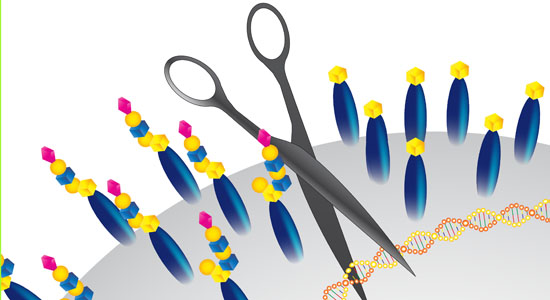
- A list of CRISPR constructs are published in our GlycoCRISPR publication (Narimatsu et al. 2018)
- Zinc Finger Nucleases
- GTf expression constructs
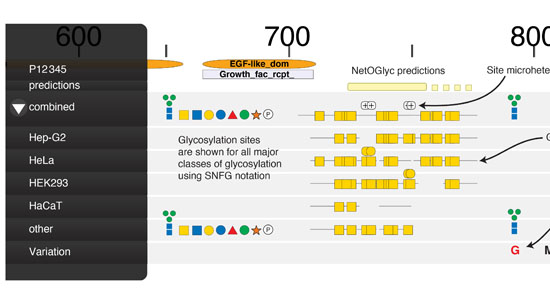
The CCG hosts informatics resources to explore the vast amounts of data produced through our glycoengineering efforts.
- Glycoproteomes: https://glycodomain.glycomics.ku.dk (Catharina Steentoft et al. 2013; Joshi et al. 2018)
- O-Glycoproteome DB: http://glycoproteomics.somee.com
- O-glycosylation predictor: https://services.healthtech.dtu.dk/service.php?NetOGlyc-4.0 (Catharina Steentoft et al. 2013)
- Open Source Software: https://github.com/CopenhagenCenterForGlycomics
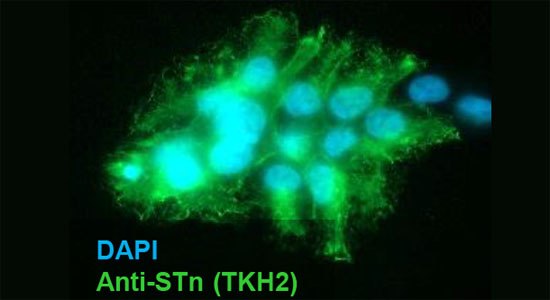
We have developed a comprehensive set of mAbs[1] for use in probing the complex biosynthesis of mucin-type O-glycosylation and the truncated O-glycans characteristically found in cancers.
The specificity of the mAbs is extensively validated in compliance with recent proposed guidelines.
A summary of these reagents is presented in this review:
A validated collection of mouse monoclonal antibodies to human glycosyltransferases functioning in mucin-type O-glycosylation.
Steentoft, Catharina, Yang, Zhang, Wang, S., Ju, T., Vester-Christensen, M. B., Festari, M. F., King-Smith, S. L., Moremen, K., Larsen, Ida Signe Bohse, Goth, Christoffer Knak, Schjoldager, Katrine Ter-Borch Gram, Hansen, Lars, Bennett, Eric Paul, Mandel, Ulla & Narimatsu, Yoshiki, 2019, I : Glycobiology. 29, 9, s. 645–65611 s.
We have also generated a series of mAbs directed to epitopes that require both glycan and peptide for reactivity. Our selection includes antibodies targeting different mucins (MUC1, MUC2, MUC4) with aberrant O-glycopeptide epitopes as well as dysadherin and fibronectin with Tn. An interesting outcome of our studies is that one of our mAbs 5E5 directed to a cancer specific epitope on mucin-1 (Tn-MUC1) is now in first phase clinical trial (CAR-T immunotherapy).
For Antibodies to human glycans. Please refer to these papers:
A Genetic Approach to Display and Dissect the Cancer-Associated O-Glycoepitome
Clausen H, Nason R, Mandel U, & Narimatsu Y, 2020, I : FASEB Journal. 34, S1, 1 s.
Glycan-directed CAR-T cells
Steentoft C, Migliorini D., King T. R., Mandel U, June C. H., & Posey A. D., 2018, I : Glycobiology. 28, 9, s. 656-669
Engineered CAR T Cells Targeting the Cancer-Associated Tn-Glycoform of the Membrane Mucin MUC1 Control Adenocarcinoma
Posey, A. D., Schwab, R. D., Boesteanu, A. C., Steentoft, Catharina, Mandel, Ulla, Engels, B., Stone, J. D., Madsen, Thomas Daugbjerg, Schreiber, K., Haines, K. M., Cogdill, A. P., Chen, T. J., Song, D., Scholler, J., Kranz, D. M., Feldman, M. D., Young, R., Keith, B., Schreiber, H., Clausen, Henrik, Johnson, L. A. & June, C. H., 21 jun. 2016, I : Immunity. 44, 6, s. 1444-1454 11 s.
A strategy for generating cancer-specific monoclonal antibodies to aberrant O-glycoproteins: identification of a novel dysadherin-Tn antibody
Steentoft C., Fuhrmann M., Battisti F., Van Coillie J., Madsen T.D., Campos D., Halim A., Vakhrushev S., Joshi H.J., Schreiber H., Mandel U., & Narimatsu Y., 2019, I : Glycobiology. 29, 4, s. 307-319 13 s.
Our collection of mAbs are available to the community.
Footnote:
[1] mAbs 5B6 (C1GalT1) and 6G8 (human GLA) are under license agreement with EMD Millipore Merck. mAbs UH3 4D8 (GalNAc-T1), UH4 4C4 (GalNAc-T2) and UH5 2D10 (GalNAc-T3) are under license agreement with Ximbio.
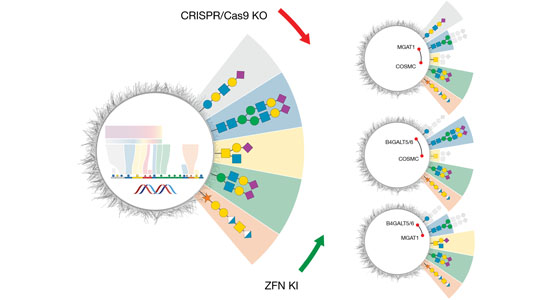
- SimpleCells
- Glycoengineered HEK293 cells
- Glycoengineered CHO cells
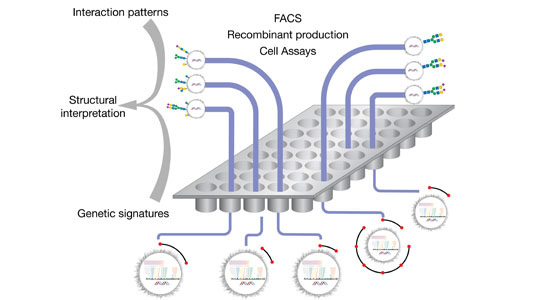
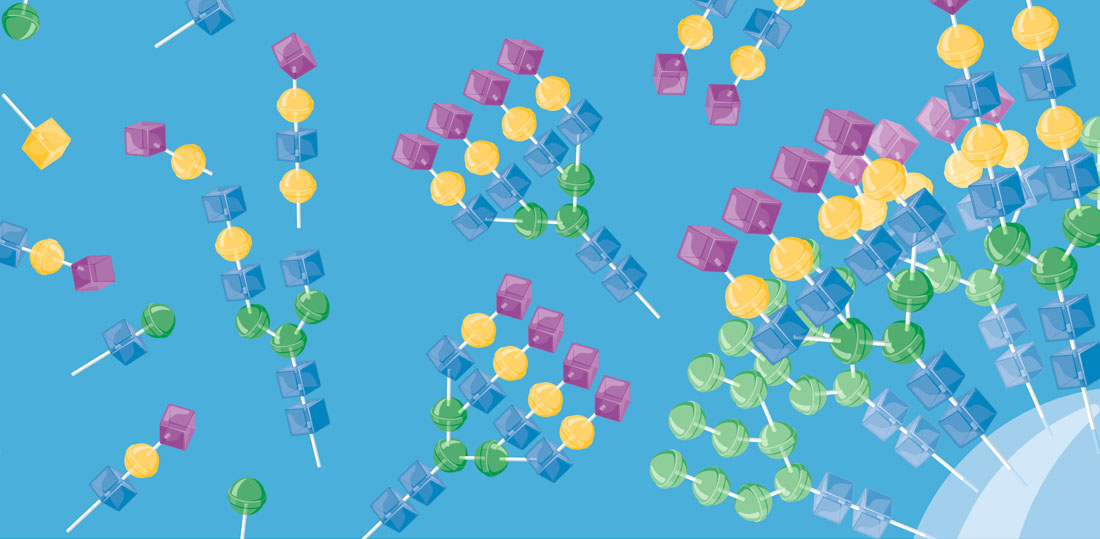
The structural diversity of glycans on cells — the glycome — is vast and complex to decipher. Printed glycan arrays displaying oligosaccharides and widely used to interrogate interactions with glycans, but these are limited resources and present glycans without the context of other glycans and glycoconjugates. To complement these we are developing cell-based glycan arrays for the human glycome including glycosaminoglycans. We use maps of glycosylation pathways (Schjoldager et al. 2020) to generate libraries of isogenic HEK293 cells with combinatorically engineered glycosylation capacities designed to display and dissect the genetic, biosynthetic, and structural basis for glycan interactions (Chen et al. 2018; Narimatsu et al. 2019). The cell-based glycan array is a self-renewable and sustainable resource we are making available to the Community (custom assaying & cell distribution).
The innovative cell-based platform is the first to display glycans in the natural context of the cell, enabling high throughput screening, discovery and dissection of distinct structure-function relationships, and production of designer therapeutic glycoproteins. The cell-based glycan arrays are useful for deciphering glycan-binding specificities of bacterial, viral and human proteins and dissection of biological roles of glycans in cell biology (Chen et al. 2018; Narimatsu et al. 2019; Büll et al. 2020). The platform also allows for the design and discovery of improved recombinant glycoprotein therapeutics (Yang et al. 2015; Schulz et al. 2018; Tian et al. 2019).
A public library of glycan images/templates and a SNFG color palette titled, “Human Glyco,” designed by Assistant Professor Hiren Joshi in Adobe Illustrator is available for use by the scientific community.
For questions regarding usage, etc., contact Hiren Joshi by email: joshi@sund.ku.dk.
References
Büll, Christian, Hiren Jitendra Joshi, Henrik Clausen, and Yoshiki Narimatsu. 2020. “Cell-Based Glycan Arrays—A Practical Guide to Dissect the Human Glycome.” STAR Protocols 1 (1): 100017. https://doi.org/10.1016/j.xpro.2020.100017.
Chen, Yen-Hsi, Yoshiki Narimatsu, Thomas M Clausen, Catarina Gomes, Richard Karlsson, Catharina Steentoft, Charlotte B Spliid, et al. 2018. “The GAGOme: A Cell-Based Library of Displayed Glycosaminoglycans.” Nature Methods 15 (11): 881–88. https://doi.org/10.1038/s41592-018-0086-z.
Joshi, Hiren J., Anja Jørgensen, Katrine T. Schjoldager, Adnan Halim, Leo A. Dworkin, Catharina Steentoft, Hans H. Wandall, Henrik Clausen, and Sergey Y. Vakhrushev. 2018. “GlycoDomainViewer: A Bioinformatics Tool for Contextual Exploration of Glycoproteomes.” Glycobiology 28 (3): 131–36. https://doi.org/10.1093/glycob/cwx104.
Narimatsu, Yoshiki, Hiren J. Joshi, Zhang Yang, Catarina Gomes, Yen Hsi Chen, Flaminia C. Lorenzetti, Sanae Furukawa, et al. 2018. “A Validated GRNA Library for CRISPR/Cas9 Targeting of the Human Glycosyltransferase Genome.” Glycobiology 28 (5): 295–305. https://doi.org/10.1093/glycob/cwx101.
Narimatsu, Yoshiki, Hiren J Joshi, Rebecca Nason, Julie Van Coillie, Richard Karlsson, Lingbo Sun, Zilu Ye, et al. 2019. “An Atlas of Human Glycosylation Pathways Enables Display of the Human Glycome by Gene Engineered Cells.” Molecular Cell 75 (2): 394-407.e5. https://doi.org/10.1016/j.molcel.2019.05.017.
Schjoldager, K.T., Y. Narimatsu, H.J. Joshi, and H. Clausen. 2020. “Global View of Human Protein Glycosylation Pathways and Functions.” Nature Reviews Molecular Cell Biology. https://doi.org/10.1038/s41580-020-00294-x.
Schulz, Morten A, Weihua Tian, Yang Mao, Julie Van Coillie, Lingbo Sun, Joachim S Larsen, Yen-Hsi Chen, et al. 2018. “Glycoengineering Design Options for IgG1 in CHO Cells Using Precise Gene Editing.” Glycobiology 28 (7): 542–49. https://doi.org/10.1093/glycob/cwy022.
Steentoft, C., M. Fuhrmann, F. Battisti, J. Van Coillie, T.D. Madsen, D. Campos, A. Halim, et al. 2019. “A Strategy for Generating Cancer-Specific Monoclonal Antibodies to Aberrant O-Glycoproteins: Identification of a Novel Dysadherin-Tn Antibody.” Glycobiology 29 (4). https://doi.org/10.1093/glycob/cwz004.
Steentoft, Catharina, Sergey Y Vakhrushev, Hiren J Joshi, Yun Kong, Malene B Vester-Christensen, Katrine T-B G Schjoldager, Kirstine Lavrsen, et al. 2013. “Precision Mapping of the Human O-GalNAc Glycoproteome through SimpleCell Technology.” The EMBO Journal 32 (10): 1478–88. https://doi.org/10.1038/emboj.2013.79.
Tian, Weihua, Zilu Ye, Shengjun Wang, Morten Alder Schulz, Julie Van Coillie, Lingbo Sun, Yen-Hsi Chen, et al. 2019. “The Glycosylation Design Space for Recombinant Lysosomal Replacement Enzymes Produced in CHO Cells.” Nature Communications 10 (1): 1785. https://doi.org/10.1038/s41467-019-09809-3.
Turnbull, Jeremy E. 2018. “Enhancing the Glycosciences Toolkit: New GAGs in the Lineup.” Nature Methods 15 (11): 867–68. https://doi.org/10.1038/s41592-018-0190-0.
Yang, Zhang, Shengjun Wang, Adnan Halim, Morten Alder Schulz, Morten Frodin, Shamim H Rahman, Malene B Vester-Christensen, et al. 2015. “Engineered CHO Cells for Production of Diverse, Homogeneous Glycoproteins.” Nature Biotechnology 33 (8): 842–44. https://doi.org/10.1038/nbt.3280.
